Nucleation¶
MICRESS® offers a number of submodels to simulate nucleation under assumption of different material and process specific conditions. Note that nucleation conditions have to be specified for each phase, individually. Without a given nucleation condition, a new phase will not appear. Nucleation conditions are always defined with respect to a certain reference phase and with respect to a certain nucleation position, e.g. the bulk of the phase or the interface to another phase. Nucleation will occur when the local undercooling evaluated with respect to the reference phase exceeds the critical undercooling required for nucleation. The critical undercooling can either be defined by the user as a fixed value or evaluated based on a specific criterion, e.g. for heterogeneous nucleation on inoculant particles.
Activation of nucleation¶
The keyword nucleation activates nucleation input. The optional parameter verbose triggers additional screen output on the number of potential and successful nucleation events. This option is very helpful to find out why a given set of nucleation parameters is not leading to the desired result, but is not recommended to be used during standard simulation. The option out_nucleation in the next line triggers additional graphical result outputs for all time-step at which a nucleus is set or a phase disappears. This option is not recommended in case of high numbers of nucleation events. In the following, the number of different nuclei types (here referred to as 'seed types') is requested.
... # Nucleation # ========== # Enable further nucleation? # Options: nucleation nucleation_symm no_nucleation [verbose|no_verbose] nucleation # Additional output for nucleation? # Options: out_nucleation no_out_nucleation no_out_nucleation # # Number of types of seeds? 1 ...
Define seed types¶
- Seed position
- Seed model
- seed growth model
- Shield effect
- Other settings, e.g. nucleation range, time check interval
Example 1¶
One seed type in the bulk
... # Input for seed type 1: # ---------------------- # Type of 'position' of the seeds? # Options: bulk region interface triple quadruple front [restrictive] bulk # Phase of new grains (integer) [unresolved|add_to_grain|split_from_grain]? 1 # Reference phase (integer) [min. and max. fraction (real)]? 0 # Which nucleation model shall be used? # Options: seed_undercooling seed_density seed_undercooling # Maximum number of new nuclei of seed type 1? # (set negative for unlimited number) -1 # Grain radius [micrometers]? 0.0000 # Choice of growth mode: # Options: stabilisation analytical_curvature stabilisation # min. undercooling [K] (>0)? 10.000 # Shield effect: # Shield time [s] [shield phase or group number] ? 10.000 # Shield distance [micrometers] [ nucleation distance [micrometers] ]? 1.000 25. # Nucleation range # min. nucleation temperature for seed type 1 [K] 0.0000 # max. nucleation temperature for seed type 1 [K] 1200.0 # Time between checks for nucleation? [s] # Options: constant from_file constant # Time interval [s] 5.0100 # Shall random noise be applied? # Options: nucleation_noise no_nucleation_noise no_nucleation_noise # ...
Seed position¶
Seeds can either be placed in the bulk, in defined regions, at interfaces, at triple or quadruple junctions or at a certain distance to a given phase (front), as illustrated in Figure 1. Note that by default, bulk or region include interfaces, interface includes triple junctions, triple junctions includes quadruple junction, and those all other high-order junctions. The optional parameter 'restrictive' allows to address pure 'bulk' without interfaces, or pure interface without triple junctions and so on. In case of region, the minimum and maximum coordinates are requested (in \rm{\mu m}) for each direction . To trigger nucleation at a predefined point, equal minimum and maximum values nay be selected. In case of front, the nuclei are placed within the reference phase, but at a certain distance to the substrate phase. This distance is specified by the user in a separate line.
Figure 1¶
Illustration of seed position options
1: interface. 2: triple junction. 3: bulk
For each seed type, the phase of the seed (nuclei) as well as the reference phase must be defined. Note that nuclei can appear only at places where the reference phase is present. In the case of concentration coupling, the driving force for nucleation is calculated as function of the local composition of this phase. In solidification simulations, the reference phase is typically the liquid phase.
In case of nucleation on interfaces or junctions, specification of an additional substrate phase is requested, see Example 2. Nucleation will only occur at interfaces where the substrate phase is present. if an additional optional second substrate phase is specified, nucleation will only occur at interfaces or junctions where both phases coexist. Both substrate phases may be defined equal to model nucleation on grain boundaries, only. The curvature of the first substrate determines the curvature contribution to the nucleation undercooling. This curvature contribution is disregarded if the substrate phase is identical to the reference phase. In the case of front, the substrate phase determines in front of which phase nucleation shall happen.
In the same line with the phase number of the new grain, two optional models can be specified, namely unresolved or add_to_grain. The option unresolved invokes a model in which fine eutectic or eutectoid phases are treated as a continuous phase mixture (see chapter Diffuse Phase Model for unresolved eutectic-or-eutectoid regions).
The option add_to_grain allows nucleation of a grain which already exists in a second place, as shown in Example 2. As the properties of the grain will be overwritten by those specified in the nucleation parameters, the option add_to_grain allows also changing properties of an existing grain. The suboptions grain_number, parent_grain or new_set must be chosen in the next line. For details see chapter Advanced Grain Manipulation.
Furthermore, the minimal and maximal fraction of the reference phase where new nuclei are allowed to appear can be optionally specified in the same line with the reference phase number. This allows the user, for example, to place new seeds at the outer part of a solid-liquid interface which increases their chances to grow before getting overgrown by the advancing solidification.
Example 2¶
Position type
interfaceand advanced optionadd_to_grain... # Type of 'position' of the seeds? # Options: bulk region interface triple quadruple front [restrictive] interface # Phase of new grains (integer) [unresolved|add_to_grain|split_from_grain]? 2 add_to_grain # Which grain number the new grains shall be added to? # Options: grain_number (number) parent_grain new_set grain_number 0 # Reference phase (integer) [min. and max. fraction (real)]? 0 0.95 1. # Substrat phase [2nd phase in interface]? 1 3 ...
Seed model¶
In case of nucleation in bulk or region, MICRESS® allows the user to choose between the standard 'seed undercooling' and the so-called 'seed density' model, which models heterogeneous nucleation on virtual particles randomly distributed in the melt.
seed_undercooling model¶
Related topics:
Example 3 demonstrates how to apply the seed undercooling model. If seed_undercooling is chosen, no explicit potential nucleation sites are predefined. The user can specify a maximum number of grains which are allowed for each seed type. If this number is exceeded, nucleation of this type stops without warning (if verbose is not activated). If a negative number is given, an unlimited number of grains are allowed to be nucleated.
Next, an explicit radius for the grains of this type has to be specified. If a value higher than the spatial discretisation \Delta x is chosen, then a grain with the corresponding size and a sharp interface is created. Note, that with concentration coupling, even if a different phase is nucleated, the new grain adopts the concentration which was present in the concerned grid cells before. Especially with TQ coupling, nucleating "big" grains will most probably lead to numerical problems and should be avoided!
If a radius value lower than the spatial discretisation \Delta x is chosen, a "small" grain consisting of only one interface cell is created. Usually, a value of 0 will be used to start with the smallest possible fraction of 2 x phMin. By this way, any kind of concentration imbalance is avoided (given that phMin has been chosen appropriately). The factor 2 given here is the default value of the so-called hysteresis factor. It can be changed manually (see section Numerical Parameters - Phase-Field Solver).
Under certain circumstances, the user may wish to specify a radius value between 0 and \Delta x. Then, a grain consisting of a single cell with a fraction of the new phase corresponding to the 3D volume will appear. Afterwards, the small grain model to be used must be specified.
Example 3¶
Further nucleation using the seed undercooling model
... # Which nucleation model shall be used? # Options: seed_undercooling seed_density [lognormal_1 | lognormal_2] seed_undercooling # Maximum number of new nuclei of seed type 3? # (set negative for unlimited number) 5000 # Grain radius [micrometers]? 0.0 ...
seed_density model¶
Related Topics
Related Examples
If seed_density option is applied in a simulation, discrete positions with discrete radii for the potential nucleation sites of the involved seed types will be determined and stored at the beginning of the simulation. During the simulation, nucleation is checked only at these predefined places. These seeding particles are not consumed by nucleation and cannot move. An exception is the moving_frame option (see section Process Conditions): If this option is selected, the predefined nucleation sites move with the frame, the sites which move out at the bottom of the domain are copied to the top line in order to keep the density of nucleation sites constant.
If seed_density is selected, the user first has to specify an integer for randomization. The random number generator has to be initialized with an arbitrary integer number (see Example 4). This ensures that, e.g., inserting a new nucleation type would not change the random positions of all other types for a given MICRESS® example.
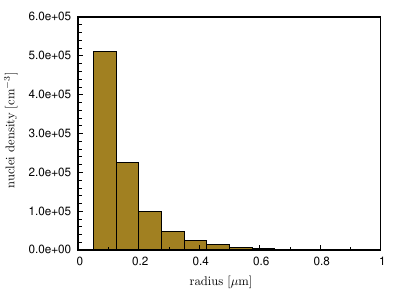
In the seed density model, the size distribution of potential nucleation sites (seeding particles) is described in terms of classes with different radii \rm{[\mu m]} and density \rm{[cm^{-3}]}, as shown in Figure 2 . For each class, the number of potential nucleation sites is calculated according to the given density and the volume of the simulation domain.
Note: In 2D simulations, the given 3D density is transformed to an equivalent 2D density which has the same average distance between the seeding particles as it would have in 3D.
Figure 2¶
Seed density classes
Using the given density, explicit positions of the potential nucleation sites are determined. The radii (and thus the local critical undercooling for nucleation on each particle) are distributed evenly according to the radius range of each seed class. Inside each class, a random radius distribution is assumed. The radius range corresponds to the radius difference to the next specified class. The finally created numbers and radius ranges for each class can be found in the .log-file.
In MICRESS®, the seed density distribution can be either defined manually by using a table form, or be generated automatically by defining a lognormal density function. In the case of table form (see Example 4), the seed classes must be specified starting with the highest radius values. At least two classes should be specified otherwise the automatically assumed radius range may lead to unexpected results.
Optionally, an additional clustering of the randomly distributed seeds can be obtained. This is done by specifying an additional cluster radius and a cluster density in the same line for each class (i.e. four real numbers in each line). The option can be helpful if experimentally a clustering of primary precipitates is observed like in some hypereutectic Al-Si alloys.
Example 4¶
Bulk nucleation using the seed density model
... # Which nucleation model shall be used? # Options: seed_undercooling seed_density [lognormal_1 | lognormal_2] seed_density # Integer for randomization? 13 # Number of seed classes? 10 # Specify radius [micrometers] and seed density [cm**-3] for class 1 0.45 1000 # Specify radius [micrometers] and seed density [cm**-3] for class 2 0.3 2000 # Specify radius [micrometers] and seed density [cm**-3] for class 3 0.25 5000 # Specify radius [micrometers] and seed density [cm**-3] for class 4 0.18 10000 # Specify radius [micrometers] and seed density [cm**-3] for class 5 0.15 20000 # Specify radius [micrometers] and seed density [cm**-3] for class 6 0.12 50000 # Specify radius [micrometers] and seed density [cm**-3] for class 7 0.10 90000 # Specify radius [micrometers] and seed density [cm**-3] for class 8 0.08 140000 # Specify radius [micrometers] and seed density [cm**-3] for class 9 0.07 250000 # Specify radius [micrometers] and seed density [cm**-3] for class 10 0.06 500000 # Determination of nuclei orientations? ...
For generating seed classes using lognormal function, either lognormal_1 or lognormal_2 option should be added next to the option seed_density. In the case of lognormal_1 (see Example 5), the user is required to give the median radius [\rm \mu m] and the standard deviation to define the lognormal density function. By lognormal_2 (see Example 6), the lognormal distribution function is determined by the most frequent radius [\rm \mu m] and an additional specific radius [\rm \mu m]. Here, the specific radius indicates a radius by a specific cumulative distribution function(CDF) value. The user is also allowed to specify the CDF value. By default, the CDF value is set to be 0.99 (or 99%), which corresponds to the maximal radius in the system. In Example 6), the value CDF=0.5 is given, then the specific radius is equivalent to the median radius. More details please refer to the section Lognormal distribution.
Example 5¶
seed density model with option lognormal_1
... # Which nucleation model shall be used? # Options: seed_undercooling seed_density [lognormal_1 | lognormal_2] seed_density lognormal_1 # Median radius in micrometer? 0.05000 # Standard deviation ? 1.00000 ...
Example 6¶
seed density model with option lognormal_2
... # Which nucleation model shall be used? # Options: seed_undercooling seed_density [lognormal_1 | lognormal_2] seed_density lognormal_2 # Most frequent radius in micrometer? 0.02000 # Specific radius in micrometer [<CDF value> (Default: 0.99)] 0.05632 0.5 ...
After the determination of lognormal density function, some parameters are needed for automatically the seed classes: the randomization integer, the number of classes, the total particle density [\rm cm^{-3}] in the simulation region, and a range of radius in which the nucleation most probably takes place.
... # Generate seed density classes: # Integer for randomization? 13 # Number of seed classes? 10 # Total seed density in cm**-3? 0.20000E+07 # Range of considered seed radius in micrometer? # <radiusMin> <radiusMax> 0.05500 0.52500 ...
Small seed treatment¶
For defining the grain growth model, the user should choose either stabilisation or analytical_curvature, and define the minimal undercooling. Details about how to choose them is described in section Microstructure.
... # Grain radius [micrometers]? 0.0000 # Choice of growth mode: # Options: stabilisation analytical_curvature stabilisation # min. undercooling [K] (>0)? 10.000 ...
If the phase of the new phase has not been set to isotropic (see Phases/anisotropy), the orientation of the new grains has to be specified. A random distribution, a fix value, an orientation range or a parent relation, i.e. a relative orientation to the grain of the local substrate phase, can be chosen. The option parent_relation applies only to nucleation at interfaces or junctions. For the option fix, the rotation angle should be given according to which relative rotation option has been chosen for the nuclei phase, see Phases/Crystallographic orientation.
... # Determination of nuclei orientations? # Options: random randomZ fix range parent_relation fix # Rotation angle? [Degree] 38.000 ...
Shield effect¶
Related Topics
Shielding means that no further grain of the same phase will be nucleated during the shield time within the shield distance of a previously nucleated grain. No categorization will be applied to this grain during the shield time (otherwise the shield properties would be lost!). The shield time is also used by the kill_metastable option (see below): No killing is performed during the shield time.
In the same line with the shield time, a shield group number can be specified. Grains with the same shield group number are shielded against each other. The default is the phase number. Note that also phase numbers can be used as shield group number. This allows for establishing shielding between different phases.
After the shield distance an optional parameter for nucleation distance can be entered. This parameter determines the minimal distance between grains of the same phase that nucleate at the same time. If no nucleation distance is given it defaults to the shield distance.
In case of the seed density model, explicit shielding is not compatible with the underlying physical model. The user is requested to specify only a shield time which still is necessary to control the categorization and kill_metastable functions.
Other settings¶
If in section Phases, categorize and anisotropic have been chosen for the nuclei phase, the user has to decide whether for this seed type categorization of the orientation values to orientation categories shall be performed. This is important, if different grains shall be assigned to the same grain number during run-time, because this is only possible if all grain properties including orientations of the grains are identical (see Phases). After the keyword categorize, the number of orientation categories can be specified, default is 36 (corresponding to 10° difference between the orientation categories in 2D).
... # Shall categorization be applied to this seed type? # Options: categorize {Number} no_categorize categorize 36 ...
For RX nucleation, the user has to specify also a minimal and maximal energy or alternatively dislocation density of the nuclei. Classically, these energies are zero. In this case, RX starts directly. If these energies differ from zero, RX simulation starts later and an initial microstructure can be built up for example in the case of RX simulation after a solidification analysis. When specifying dislocation densities, their value must be very small and significantly lower than the specified dislocation threshold.
Further, MICRESS® requests a minimum and a maximum nucleation temperature for the actual seed type. A minimum and maximum temperature should be chosen around the temperature where nucleation is expected. The parameters primarily help to minimize unnecessary nucleation checks and thus to improve performance. In some cases with TQ coupling, checking nucleation too far from the temperature where the new phase gets stable can cause numerical problems. Then, proper values have to be found, depending on the system. In a first trial, it is wise not to restrict the nucleation range.
The time between checks for nucleation determines the frequency of nucleation checking. If chosen too high, insufficient seeding may occur in spite of high local undercooling. If chosen too small, an unnecessarily high numerical effort and a corresponding performance loss can be the consequence. Note that in case of TQ coupling, each check for nucleation requires time-consuming calls to the TQ interface for each checked position. A possible performance loss can be monitored using the .TabP or .TabTQ output. Time-dependent checking frequencies can be specified by input from file.
... # Nucleation range # min. nucleation temperature for seed type 1 [K] 0.00 # max. nucleation temperature for seed type 1 [K] 2000.00 # Time between checks for nucleation? [s] # Options: constant from_file constant # Time interval [s] 0.10 ...
Noise can be added to the nucleation undercooling. In that case, the amplitude for the noise level and a seed for the random number generator will be asked later (see below).
The noise is applied as
where k is user defined noise amplitude, 0 \le {\rm random} \le 1 is the value of the random number generator and \Delta G is the nucleation driving force.
... # Shall random noise be applied? # Options: nucleation_noise no_nucleation_noise nucleation_noise # Factor for random noise? # (applied as DeltaT -> (1+Factor*(RAND-1/2))*Delta 0.1 ...
Input for all seed types¶
... # Max. number of simultaneous nucleations? # ---------------------------------------- # (set to 0 for automatic) 0 # # Shall metastable small seeds be killed? # Options: kill_metastable no_kill_metastable no_kill_metastable ...
After specification of all seed types, some few inputs remain to be done which apply to all seed types. First of all, if any of the seed types is using random noise, an integer number for randomization has to be given to assure reproducibility. The maximum number of simultaneous nucleation events is the number of grains of all seed types allowed to be nucleated in the same time step. A list of possible seeds is created and ordered with respect to the undercooling or driving force for each nucleus. If the maximum number is exceeded, the less favourable seeds are discarded. The option may lead to unexpected results if more than one seed type is defined and therefore should be used carefully. Setting the maximum number of simultaneous nucleation events to zero (i.e. automatic) removes this limit.
In some cases, "stabilised" small grains can erroneously reach a metastable state at which they stop growing, but also do not vanish because their stabilisation implies a reduced curvature. By enabling the flag kill_metastable, the stabilisation of small grains is removed after their shield time has elapsed. This makes sure that "metastable grains" can vanish correctly. The option kill_metastable is also relevant in case of categorization, because after "killing", small stabilized grains are considered as "big" and such can be assigned to a common grain number.
Important: The kill_metastable flag is only relevant for small grains which use the stabilisation model.